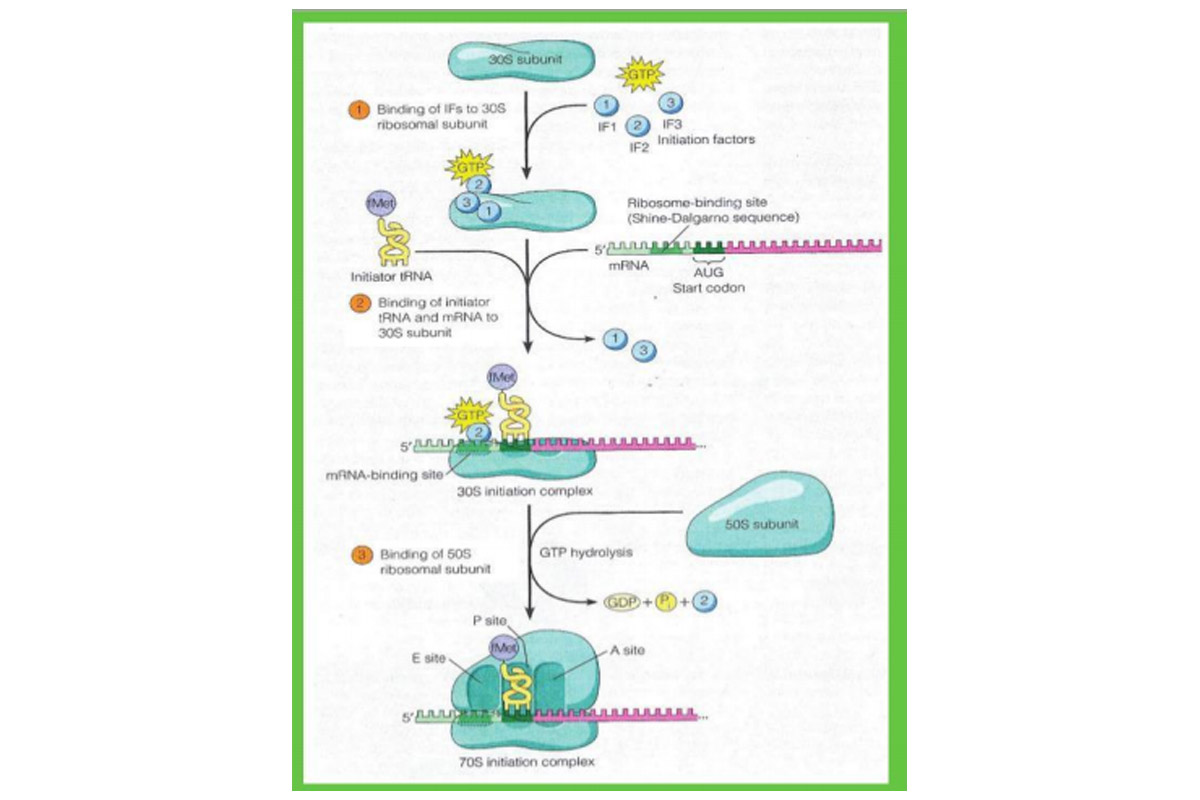
The initiation of translation in prokaryotes reveals that initiation can be subdivided into three distinct steps. In step (1), three initiation factors – called IF1, IF2, and IF3 – bind to the small (30S) ribosomal subunit, with GTP attaching to IF2.
In step (2) mRNA and the tRNA carrying the first amino acid bind to the 30S ribosomal subunit. The mRNA is bound to the 30S subunit in its proper orientation by means of a special nucleotide sequence called the mRNA’s ribosomebinding site (also known as the Shine-Dalgamo sequence, after its discoverers). This sequence consists of a stretch of three to nine purine nucleotides located slightly upstream of the initiation codon. These purines in the mRNA form complementary base pairs with a pyrimidinerich sequence at the 3’ end of 16S rRNA, which forms the ribosome’s mRNA-binding site.
The importance of the mRNA-binding site has been shown by studies involving colicins, which are proteins produced by certain strains of Escherichia coli that can kill other types of bacteria. One such protein, colicin E3, kills bacteria by destroying their ability to synthesise proteins. Upon entrance into the cytoplasm of susceptible bacteria, colicin E3 catalyses the removal of a 49-nucleotide fragment from the 3’ end of 16S rRNA, destroying the mRNA-binding site and thereby creating ribosomes that can no longer initiate polypeptide synthesis.
Advertisement
The binding of mRNA to the mRNA-binding site of the small ribosomal subunit places the mRNA’s AUG start codon at the ribosome’s P site, where it can then bind to the anticodon of the appropriate tRNA. The first clue that a special kind of tRNA is involved in this step emerged when it was discovered that roughly half the proteins in E coli contain methionine at their N-terminal ends. This was surprising because methionine is a relatively uncommon amino acid, accounting for no more than a few percent of the amino acids in bacterial proteins.
The explanation for such a pattern became apparent when it was discovered that bacterial cells contain two different methionine-specific tRNAs. One, designated tRNAMet, carries a normal methio-nine destined for insertion into the internal regions of polypeptide chains. The other, called tRNAfMet,, carries a methionine that is converted to the derivative N-formylmethionine (fMet) after linkage to the tRNA. In Nformylmethionine, the amino group of methionine is blocked by the addition of a formyl group and so cannot form a peptide bond with another amino acid; only the carboxyl group is available for bonding to another amino acid. Hence N-formylmethionine can be situated only at the N-terminal end of a polypeptide chain, suggesting that tRNAfMet functions as an initiator tRNA that starts the process of translation.
The idea was soon confirmed by the discovery that bacterial polypeptide chains in the early stages of synthesis always contain Nformylmethionine at their N-terminus. Following completion of the polypeptide chain (and in some cases while it is still being synthesised), the formyl group, and often the methionine itself, is enzymatically removed.
During initiation, the initiator tRNA with its attached N-formylmethionine is bound to the P site of the 30S ribosomal subunit by the action of initiation factor IF2 (bound to GTP), which can distinguish initiator tRNA^61 from other kinds of tRNA. This attribute of IF2 helps to explain why AUG start codons bind to the initiator tRNA0461, whereas AUG codons located elsewhere in mRNA bind to the non-initiating tRNAMet. Once tRNAtMet enters the P site, its anticodon basepairs with the AUG start codon in the mRNA, and IF1 and IF3 are released. At this point, the 30S subunit with its associated IF2-GTP, mRNA, and N-formylmethionyl tRNA0461 is referred to the 30S initiation complex.
Finally, in step (3), the 30S initiate complex is joined by a free 50S ribosomal subunit, generating the 70S initiation complex. Binding of the 50S subunit is driven by hydrolysis of the GTP associated with IF2, which occurs as IF2 leaves the ribosome. At this stage, all three initiation factors have been released.
The initiation process in eukaryotes involves a different set of initiation factors (called elFs), a somewhat different pathway for assembling the initiation complex, and a special initiator tRNAMet that – like the normal tRNA for methionine but unlike the initiator tRNA of prokaryotes – carries methionine that does not become formylated. The initiation factor eIF2, with GTP already attached, binds to the initiator methionyl tRNAMet before the tRNA then binds to the small ribosomal subunit along with other initiation factors. The resulting complex next binds to the 5’ end of an mRNA, recognising the 5’ cap (in some situations the complex may instead bind to an internal ribosome entry sequence or IRES, which lies directly upstream of the start codon of certain types of mRNA, including viral mRNAs).
After binding to an mRNA, the small ribosomal subunit, with the initiator tRNA in tow, scans along the mRNA and usually begins translation at the first AUG triplet it encounters. The nucleotides on either side of the eukaryotic start codon seem to be involved in its recognition; ACCAUGG (also called a Kozak sequence) is a common start sequence, where the underlined triplet is the actual start codon. When the initiator tRNAMet base-pairs with the start codon, the large ribosomal subunit joins the complex.
The writer is associate professor and head, department of botany, Ananda Mohan College, Kolkata
Advertisement