US scientists develop new CRISPR toolkit to allow remote-controlled genome editing
US scientists have developed a new CRISPR toolkit to boost the treatment of genetic disorders.
Here’s a look at the process of DNA renaturation that led to the discovery of repeated sequences in DNA.
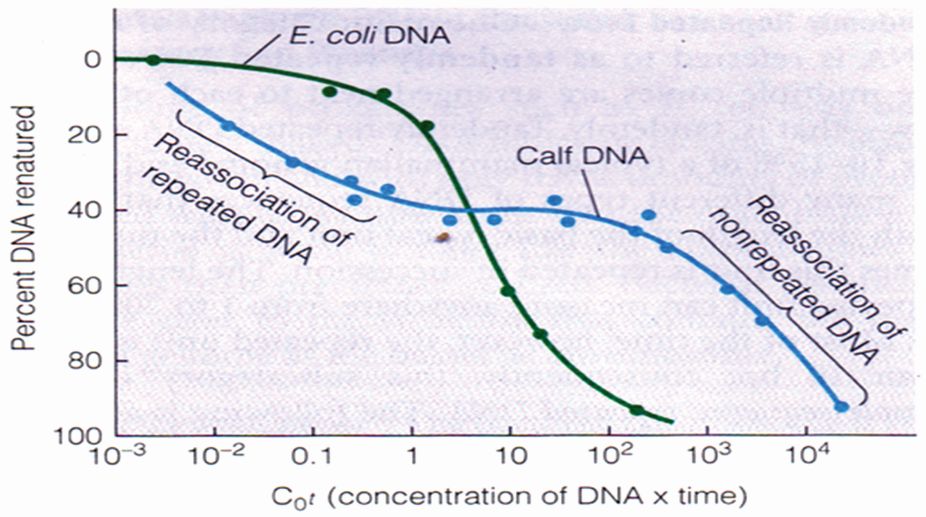
The calf DNA that reassociates more rapidly than the bacterial DNA consists of repeated sequences.
The enormous size of the human genome raises a more fundamental question — is the large amount of DNA found in the cells of humans and other higher eukaryotes simply a reflection of the need for thousands of times more genes than are present in bacterial cells, or are other factors at play as well? A major breakthrough in answering this question occurred in the late 1960s, when DNA renaturation studies carried out by Roy Britten and David Kohne led to the discovery of repeated sequences in DNA.
In the Britten and Kohne experiments, DNA was broken into small fragments and dissociated into single strands by heating. The temperature was then lowered to permit the single-stranded fragments to renature.
Advertisement
The rate of renaturation depends on the concentration of each individual kind of DNA sequence; the higher the concentration of any given kind of DNA sequence in the sample, the greater the probability that it will randomly collide with a complementary strand with which it can reassociate.
Advertisement
How would the renaturation properties of different kinds of DNA be expected to compare? As an example, let us consider DNA derived from a bacterial cell and from a typical mammalian cell containing a thousand-fold more DNA. If this difference in DNA content reflects a thousand-fold difference in the kinds of DNA sequences present, then bacterial DNA should renature 1,000 times faster than mammalian DNA.
The rationale underlying this prediction is that any particular DNA sequence should be present in a thousand-fold lower concentration in the mammalian DNA sample because there are a thousand times more kinds of sequences present, and so each individual sequence represents a smaller fraction of the total population of sequences.
When studies comparing the renaturation rates of mammalian and bacterial DNA were actually performed by Britten and Kohne, the results were not exactly as expected.
The data obtained for calf and E. coli DNA renaturation is plotted as a function of the starting DNA concentration multiplied by the elapsed time. This parameter of DNA concentration X time, or C0r, is employed in place of time alone because it permits direct comparison of data obtained from reactions run at different DNA concentrations.
When graphed this way, the data reveal that calf DNA consists of two classes of sequences that renature at markedly different rates. One type of sequence, which accounts for about 40 per cent of the calf DNA, renatures more rapidly — at a lower C0t value — than bacterial DNA.
The most straightforward explanation for this unexpected result is that calf DNA contains repeated DNA sequences that are present in multiple copies. The existence of multiple copies increases the relative concentration of such sequences, thereby generating more collisions and a faster rate of re-association than would be expected if each sequence were present in only a single copy.
The remaining 60 per cent of the calf DNA renatures about a thousand times more slowly than E. coli DNA, which is the behaviour expected of sequences present as single copies. This fraction is therefore called non-repeated DNA to distinguish it from the repeated sequences that renature more quickly.
Non-repeated DNA sequences are each present in one copy per genome. Most protein-coding genes consist of non-repeated DNA, although this does not mean that all non-repeated DNA codes for proteins.
In bacterial cells virtually all the DNA is non-repeated, whereas eukaryotes exhibit a large variation in the relative proportion of repeated and non-repeated sequences. This provides an explanation, at least in part, for the mystery of the seemingly excess amount of DNA in species such as Trillium.
This organism contains a relatively high proportion of repeated DNA sequences. Using the sequencing techniques described earlier, researchers have been able to determine the base sequences of various types of repeated DNAs and to classify them into two main categories — tandemly repeated DNA and interspersed repeated DNA.
One major category of repeated DNA is referred to as tandemly repeated DNA because the multiple copies are arranged next to each other in a row — that is, tandemly. Tandemly repeated DNA accounts for 10 to 15 percent of a typical mammalian genome and consists of many different types of DNA sequences that vary in both the length of the basic repeat unit and the number of times this unit is repeated in succession. The length of the repeated unit can measure anywhere from one to 2,000 bp or so.
The second main category of repeated sequence DNA is interspersed repeated DNA. Rather than being clustered in tandem arrangements, the repeated units of this type of DNA are scattered around the genome.
A single unit tends to be hundreds or even thousands of base pairs long and its dispersed “copies”, which may number in the hundreds of thousands, are similar but usually not identical to each other. Interspersed repeated DNA makes up 25 to 40 percent of most mammalian genomes.
The writer is associate professor, head, department Of botany, Ananda Mohan
college, Kolkata, and also Fellow, botanical society of Bengal, and can be contacted at tapanmaitra59@yahoo.co.in
Advertisement